The ITHANET Portal is a website dedicated to thalassaemia and other haemoglobinopathies. The portal provides:
- the latest news, events, publications and clinical trials in the field,
- easy access to international communities, organisations and experts concerned with haemoglobinopathies,
- techniques, protocols, guidelines and information on haemoglobinopathies and
- free access to databases and tools relevant to different aspects of haemoglobinopathies, such as sequence variations, epidemiology and phenotype.
Registered users are automatically signed to the ITHANET Newsletter list and, thus, are able to receive to the inbox the most recent updates and announcements of the ITHANET portal.
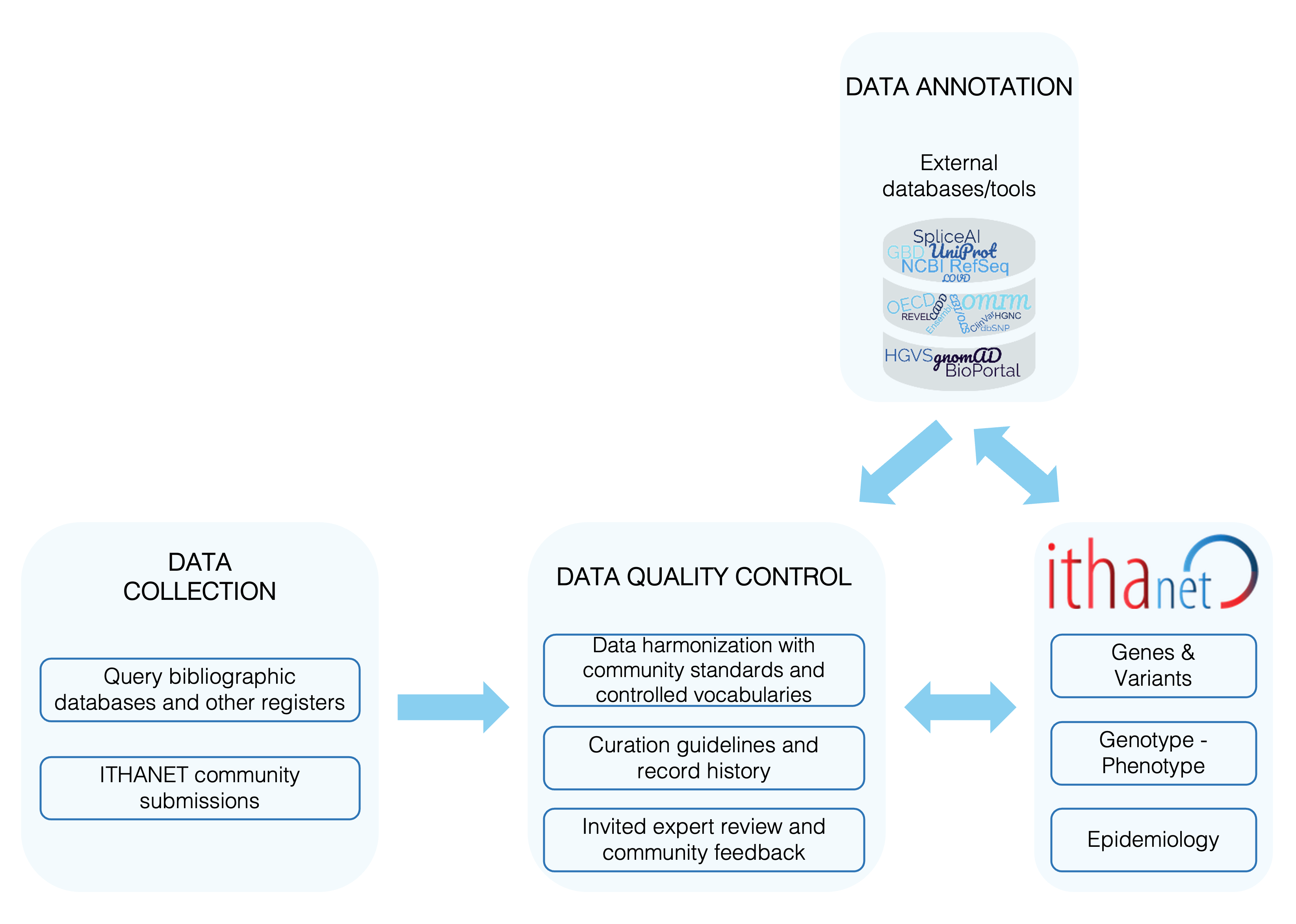
ITHANET curators adhere to internally developed operating procedures to guide curation of ITHANET databases using evidence obtained from peer-reviewed publications, meeting abstracts and direct submissions to ITHANET. These curation guidelines promote standardization of data across ITHANET and build on standard operating procedures and vocabularies to improve data quality control and interoperability.
The identification of data primarily depends on regular monitoring of literature databases (PubMed) via automatic weekly searches for hemoglobinopathy-specific keywords, as described in Kountouris et al.. The full text and supplementary material of each paper are read and evaluated by an ITHANET curator, and data are manually extracted, formatted and standardized in line with internal procedures. All information added to an entry during the manual curation process is linked to its original source, while direct submissions are given microattribution thus enabling users to trace the origin of each piece of information and evaluate it. Entries are periodically subjected to a series of semi-automated checks to maintain the quality and accuracy of the data.
IthaGenes database comprises the Gene and the Variant knowledge models, which consist of structured data and a free text field to provide evidence summaries. Features are automatically generated or embedded in the interface, whenever possible. For a Gene record to be created, it must be associated with at least one disease- or trait-associated variant and/or gene-level experimental evidence (Strande et al.). The official gene name must comefrom the HGNC in addition to listing other gene names and symbols reported in literatureand other databases. Other information includes gene biotypes (Ensembl annotation), coordinates (RefSeq) and external links to databases(NCBI, HGNC, OMIM, UniProt, PDB). For a Variant record to be created, it must be supported by evidence for linkage or association with adisease or trait and be linked to an existing Gene record. Curatable entities include, among others,HGVS descriptions, variant identifiers and position from databases (dbSNP, ClinVar,UniProt, HbVar), variant type and consequence mapped to Sequence Ontology, phenotypicdescriptions linked with terms of the HPO, pathogenicity predictions(Tamana et al.), and relevant citations.
IthaChrom and IthaCNVs tools curate elements of variant-level data and each new entry must be linked with the respective Variant record. IthaChrom provides high-performance liquid chromatography (HPLC) sample images for variant haemoglobins that are linked-out to the original source, provided by BioRad Inc. IthaCNVs reports copy number variations (CNVs) with information on specific MLPA probes (as described in the MRC Holland documentation), coordinates, breakpoints, and affected globin gene(s) and/or regulatory elements, as annotated in IthaGenes (Minaidou et al.).
IthaPhen database comprises mainly fields that contain data in a predefined format. An IthaPhen record can be created if it includes the genotype (provided all involved variants are recorded in IthaGenes) and at least one phenotypic data element. Information includes phenotypes relevant to haemoglobinopathies, demographics and family segregation data, and can only be added if linked to a source element (e.g., publication or ITHANET contributor). Critical limits of laboratory data are obtained from published best practice guidelines (EMQN, NHS trust documentation) and are also part of the internal quality control procedure.
IthaMaps database provides country-level and region-specific data. IthaMaps records already exist for each country and can includeinformation on a pre-defined set of parameters (healthcare policies, prevalence, incidence and variant allele frequencies in theglobin genes) as they become available. Sources are documented for each parameter and for each country entry. Each parameter ismanually reviewed periodically to include new or updated data. Country experts with links to ITHANET are invited to review data foreach completed entry. Migration and Global Burden of Disease data are automatically retrieved for each country and linked via theGBD and OECD databases, respectively.
Community engagement is an integral part of the ITHANET curation procedure. Data is reported in a pre-defined, standardized formatand all information added in ITHANET is supported by microattribution. . Contributors must provide details for their affiliation andan expert profile is created listing all their contributions in the different ITHANET databases.All records in ITHANET can be edited or deleted. Changes in ITHANET records are kept in an internal change log in the database.
Please use the Contact Form to ask any questions or make any comment for the ITHANET Portal.
Please click on 'News' under the option 'Latest information' of the main menu to see a list of the most recent news added in ITHANET.
You can click on the title of the item of your interest to read more information or to share it to social networks.
Please click on 'Events' under the option 'Latest information' of the main menu to see a list of all future events related to haemoglobinopathies.
You can use the 'Search' box to perform live search in the table, while you can find more information by clicking on the title and navigate to the official website of the event.
Finally, we also keep an archive of past events, which can be found by clicking on the link below the events table.
Please click on 'Publications' under the option 'Latest information' of the main menu to see the list of publications related to haemoglobinopathies.
This list is updated every week.
You can use the 'Search' box to find publications of your interest, while you can navigate to an article's PubMed page by clicking on the publication title.
You can see the list of clinical trials related to haemoglobinopathies by navigating to 'Clinical Trials' under the option 'Latest information' of the main menu.
The list of clinical trials is updated every week and retrieved from www.clinicaltrials.gov.
You can use the Search' box to perform live search in the table, while you can navigate to the trials description page at www.clinicaltrials.gov by clicking on the title.
Please use the 'Community' option of the main menu to see thelist of organisations and experts related to haemoglobinopathies.
Each organisations is assigned to one or more types based on its activities. By clicking on the organisation's name, you can see detailed information aboutthe organisation and navigate to its website for more information.
Similarly, the expertise of each expert is reported in the 'Experts' table, with more detailed description and contact details available by clickingeach expert's name.
ITHANET is supported by the European Union through FP6 and FP7 research projects, the Cyprus Research Promotion Foundation and the Cyprus Institute of Neurology and Genetics.
IthaGenes is an interactive archive of all sequence variations affectinghaemoglobinopathies, including the globin loci and disease modifiers, such as BCL11A and KLF1.
Apart from the genes and mutations, IthaGenes stores and organizes phenotype, epidemiology, HPLC data, as well as related publications and external links, while embeddingthe NCBI sequence viewer in the website for detailed graphical representation of each variation and gene.
Ithagenes provides information about genes and variations related to haemoglobin disorders.
In addition, IthaGenes stores and organizes phenotype,epidemiology, HPLC data, as well as related publications and external links, while embedding the NCBI sequence viewer in the website for detailed graphical representation of each variation and gene.
"IthaGenes" is curated by The IthaGenes Curation Team, which includes the following scientists:
- Dr. Petros Kountouris
- Dr. Carsten W Lederer
- Dr. Coralea Stephanou
- Dr. John Old
- Dr. Marina Kleanthous
The Committee Members:
- Erol Baysal
- Alex Felice
- Jan Traeger Synodinos
- Ali Taher
- Maria Sitarou
- Yeshim Aydinok
- Roberto Gambari
- Swee Lay Thein
- Suthat Fucharoean
- Stefano Rivella
- Lila Yannaki
- Cornelis L. Hartveld
- Petr Holub
- John Old
- Michael Angastiniotis
- Marina Kleanthous
- Jim Vadolas
- Aurelio Maggio
- Soteroulla Christou
IthaGenes receives weekly updates of the most recent publications from PubMed using the following search query:
'thalassemia[tiab] OR thalassaemia [tiab] OR hemoglobin [tiab] OR haemoglobin [tiab] ORsickle-cell [tiab] OR hemoglobinopathies [tiab] OR haemoglobinopathies[tiab]'.
The list of publications is manually filtered by the IthaGenesCuration Team to identify new variations and epidemiological information about haemoglobinopathies.
The new data are subsequently added into the database.
In IthaGenes, you have three different search options:
- Live Search Box: You can use the 'Search' box at the top of the table listing all IthaGenes tables (Gene list, Mutation list and References list).This provide very fast live search based on the text you type in the search box. Using this search option, you can only search the columns that are shown any given time in the table.
- Quick Filtering: A second option is using the Quick Filtering options that can be found in the menu over the Mutations table.These options are predefined searches in order to make common searcheseasily accessible and fast and eliminate the need to use the more complex Advanced Search form (see below)
- Advanced Search: If you cannot find the required information about particular variations using the above two options, we provide an Advanced Search option, where you can search for every field in the database using any combination.
Please click of the row showing the IthaGenes entry of your interest to navigateto the detailed description of the variant.
Each IthaGenes entry is assigned to one functional category:
- Globin-gene causative mutations are mutations located at the globin loci and cause a haemoglobinpathy
- Disease modifying mutations are mutations located on other loci of the genome and cause a particular phenotype (not necessarily a haemoglobinpathy). There entries may also have a trans-acting effect on genes related to haemoglobinopathies.
- Neutral polymorphisms include polymorphisms that do not have any effect on the phenotype or may have a trans-acting effect on the genes related to haemoglobinopathies
Please use the 'Print' button at the top left of the table.
This will take launch your browser's print options and you can print the informationcurrently displayed on the table (in case you have filtered/searched the initialdata).
The Sequence Viewer is a tool provided by the NCBI and is embedded in IthaGenes.
On the viewer, you can see the location of each mutation and find neighbouringmutations that may be of interest.
For more information on the use of the NCBI sequence viewer, please visit the official website.
Please note that the Sequence Viewer is not installed on ITHANET servers, but it is embedded in the page using javascript.Therefore, ITHANET is not responsible for occasional unavailability of the viewer.
When available, IthaGenes provides external links to HbVar,NCBI dbSNP, OMIM,ClinVar and SwissVar for each variation.
For each gene, we provide links to NCBI Gene,NCBI GenBank, HGNC,UniProt, OMIMand PDB.
For any suggestions about additional external links please use the >Contact Form.
Please use the Contact Form to report any correction or new content for IthaGenes.
If you are referring to a specific IthaGenes mutation, please provide the IthaID in your comments.
IthaMaps is a database that stores and organises informationabout the epidemiology of haemoglobinopathies and illustrates and compares thedata on a map.
IthaMaps provides the following information:
- Information of haemoglobinopathy-related healthcare policies in each country, including screening and preventionprogrammes, existence and availability of electronic parient registries anddedicated treament centres etc.
- Information on the status of major haemoglobinopathies ineach country, including prevalence and incidence of thalassaemias and sickle-cell disease.
- Relative allele frequencies of specific mutations in eachcountry, with detailed information about each study (sample size, ethnicgroups, specific geographical areas etc.). Where possible, a more detailedregion-specific presentation of the frequencies is provided within thecountry.
"IthaMaps" is curated by The IthaMaps Curation Team, which includes the following scientists:
- Dr. Petros Kountouris
- Dr. Carsten W Lederer
- Dr. Coralea Stephanou
- Dr. John Old
- Dr. Marina Kleanthous
The Committee Members:
- Erol Baysal
- Alex Felice
- Jan Traeger Synodinos
- Ali Taher
- Maria Sitarou
- Yeshim Aydinok
- Roberto Gambari
- Swee Lay Thein
- Suthat Fucharoean
- Stefano Rivella
- Lila Yannaki
- Cornelis L. Hartveld
- Petr Holub
- John Old
- Michael Angastiniotis
- Marina Kleanthous
- Jim Vadolas
- Aurelio Maggio
- Soteroulla Christou
Like IthaGenes, the updating of IthaMaps is basedon published literature the curation team receives every week from PubMed using the following search query:
'thalassemia[tiab] OR thalassaemia [tiab] OR hemoglobin [tiab] OR haemoglobin [tiab]OR sickle-cell [tiab] OR hemoglobinopathies [tiab] OR haemoglobinopathies [tiab]'.
The list of publications is manually filtered by the IthaMapsCuration Team to identify new epidemiological information about haemoglobinopathies.
The new data are subsequently added into the database.
Information on healthcare policiesare not always available in published articles, so further annotation is achievedwith personal communication with local experts and from informationfrom presentations and reports.
A search form is available at the top of IthaMaps page.
The user can search for a specific country or for informationabout healthcare policies, haemoglobinopathy status and relative frequencies of specific mutations.
Additional options appear based on the selection: Healthcare policies and major haemoglobinopathies status are available for the world maponly, while country selection and specific mutation frequencies can be combined. In the latter case, region-specific information are displayed on the countrymap, if available.
Information on the frequencies of specific muations can be customised based onthe selection of country- or region-specific representation, sample size and the type of statistics to be used for the illustration (average values, minimum or maximum).
IthaMaps uses the HighMaps javascript libraryfor displaying the epidemiological information, which provides exporting functionality.
The resulting map after any custom search can be exported in a variety of formatsand be used for your presentations or for other purposes.
To export a map, you can use the button at the top right of the map.
Please use the Contact Form to report any correction or new content for IthaMaps.
IthaPhen is a searchable and intuitive database of public anonymised case reportsand it integrates genotypic data with haematological, biochemical, histological, and clinical data.
IthaPhen consists of primary case level data and aggregated, visualised reportsper queried genotype. IthaPhen integrates genotypic data with haematological,biochemical, histological, and clinical data.
Who is the curator of IthaPhen?
IthaPhen allows search by specific allele and/or by allele phenotype.
When querying the database for specific allele, first select the specific variantfilter, then the locus it resides and the variant from the dropdown variant list.
If you would like to query the database for individuals with variants classifiedunder an IthaGenes allele phenotype group, select the allelephenotype filter and then one or multiple allele phenotypes.Follow the same steps for the second allele.Heterozygosity can be declared with the wild type filter in Allele 2.You can query complex genotypes by adding more rows and providing filters for the other loci.
If your search does not match any archived data, please advise Relevant Searches tab for queries involving variants in your primary search.If searching for a combination of variant and globin phenotype, the latter isignored in relevant searches.
Individual case data can be viewed by following the patient ID link from all tabsspecific for a category of parameters.
If the study did not test for all possible variants common for the ethnic groupof the individual, then the information was kept with a specific mark – 'unknown'— to record the information whilst also denoting that it is not complete.
Phenotypic effects which cannot be quantified and are annotated with categoricalvalues are often subjected to different classification systems in different publications.
For example, depending on the research group the result of a similar histological experiment for anaemia could be reported as “marked” or “moderate”.
Therefore, the number of qualitative categories used in IthaPhen was reduced to those most commonly used by the community and less common ones weremapped to this set after careful curation e.g. when one publication reported “marked”anaemia and another “moderate” anaemia, we used the term/annotation “moderate”for both these terms.
In cases where haemoglobin was measured by different methods for the same individual,we keep both measurements for the individuals record but use only the measurementwith the most trusted method for calculating the summary statistics of the genotype.You can find the list of methods by priority order, below:
- High-performance liquid chromatography (HPLC)
- Capillary electrophoresis (CE)
- Hb Electrophoresis (E)
- Chromatographic profile (ChP)
- Alkali denaturation (AD)
- Column chromatography (CC)
- Spectrophotometry (S)
- Elution electrophoresis (EE)
- Monoclonal antibody (MA)
- Betke method (BM)
- Isoelectrofocusing (IEF)
- Immunofluorescence (IF)
- Mass spectrometry (MS)
- Microchromatofocusing (M)
- Denaturing Gradient Gel Electrophoresis (DGGE)
- DEAE- Sephadex chromatography (DSC)
- Microchromatography (MG)
IthaPhen accepts direct contributions of unpublished data.Please complete the submission form at the following link.If you are providing case data of individuals with novel variants,please also complete section B of the IthaGenes submission form. In any other case, please provide the relevant IthaID when providing the individual's genotype
"The Offspring PND/PGD recommendation tab on IthaPhen provides recommendationsabout prenatal or preimplantation genetic diagnosis, depending on the user-definedgenotype query, as per the EMQN guidelines.'
